If you want to skip the background for now, go and download the data directly.
This year’s contest deals with the simulation of activity within the human brain, especially the changes within its network structure. Current understanding suggests, that a large amount of connections between neurons via synapses is constantly changing, leading to alteration of brain functionality. This includes positive effects such as the creation of memories [ 1 ]
or negative ones such as limitations caused by accidents or illnesses [ 2 ]
.
To understand these effects on a neuronal level, scientists hope to artificially recreate networks that work similarly to the human brain with the help of simulation models. The data provided in this contest is based on clinical measurements of actual human brains as well as the Model of Structural Plasticity by Butz & Van Ooyen [ 2 ]
which can be coarsely summarized as follows:
The brain is modeled by a network of neurons.
Each neuron is represented by a node with a number of properties such as location, calcium concentration, and target calcium concentration.
It can further carry axonal (“plugs”) and dendritic (“sockets”) elements.
If two neurons are spatially close and exhibit unused complementary elements, they have a probability to form a synapse, which is represented by a connection in the network through which electric energy can be passed from one neuron to another if it “fires” based on external input and the Izhikevich model [ 3 ]
.
When a neuron fires, its calcium level increases only to decrease again over time.
In this model, neurons aim to reach a certain target calcium concentration based on their current calcium level. To achieve this, it either seeks to increase or decrease the level of incoming electrical activity resulting in the addition or removal of elements, which, in turn, leads to the formation, rewiring, or deletion of synapses.
Thus, in each simulation step, each neuron calculates its energy income and calcium level based on this model, followed by the addition or deletion of elements, leading to changes in the network structure. This can be repeated until the system has reached a stable configuration or stopped on demand.
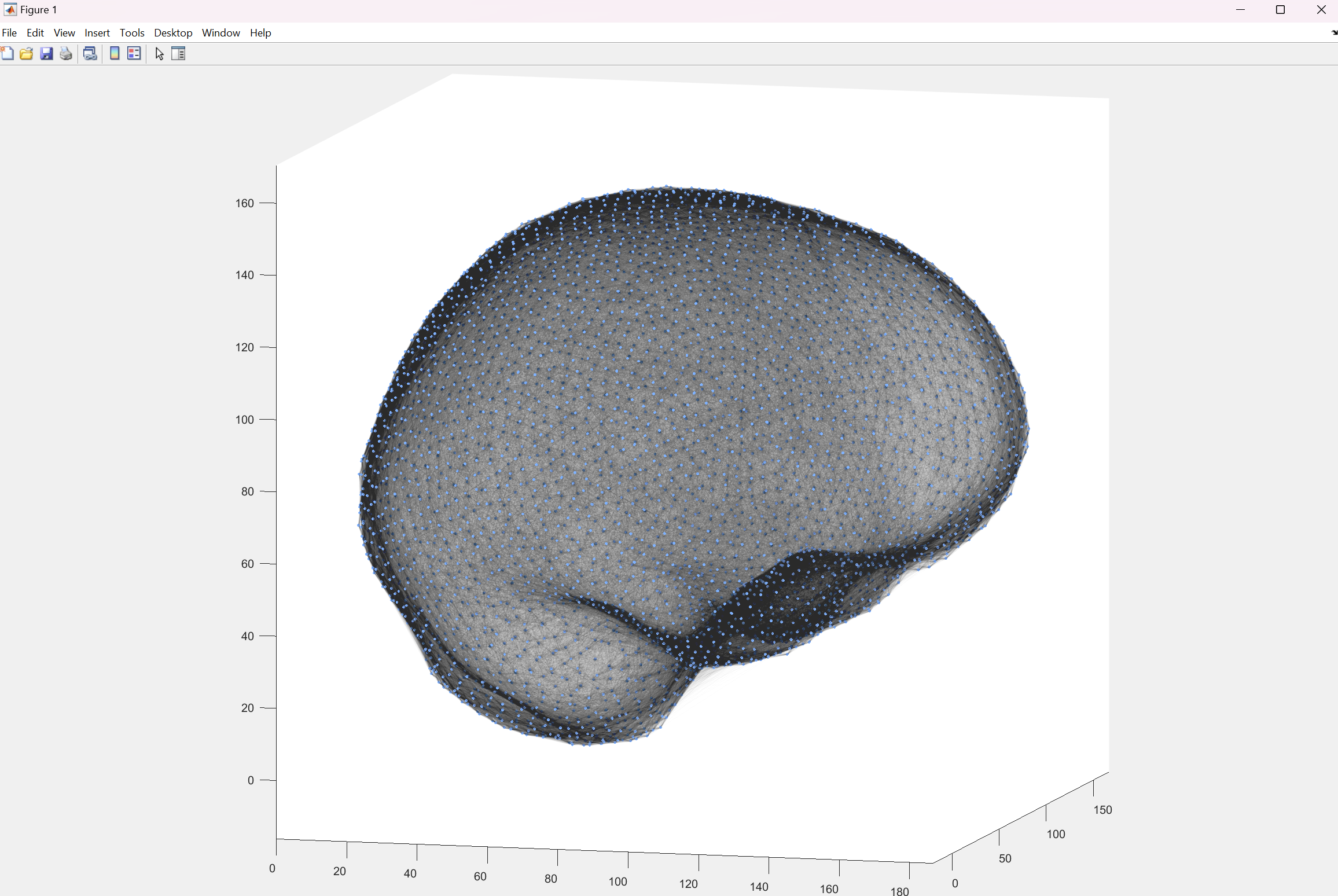
The following animation shows a simple visualization of a similar dataset: The neurons are represented as spheres at the appropriate locations and colored by their current calcium level.
On the front page you can also see a visualization containing line renderings of ingoing connections (synapses) of the same dataset.
Visualization can help to understand these processes (see, e.g., [ 4 ] ). Therefore, the goal of this contest is to enable domain scientists as well as simulation scientists to better evaluate their models, compare, and explore the simulation results. Based on this, we have provided a list of visualization tasks for this contest.